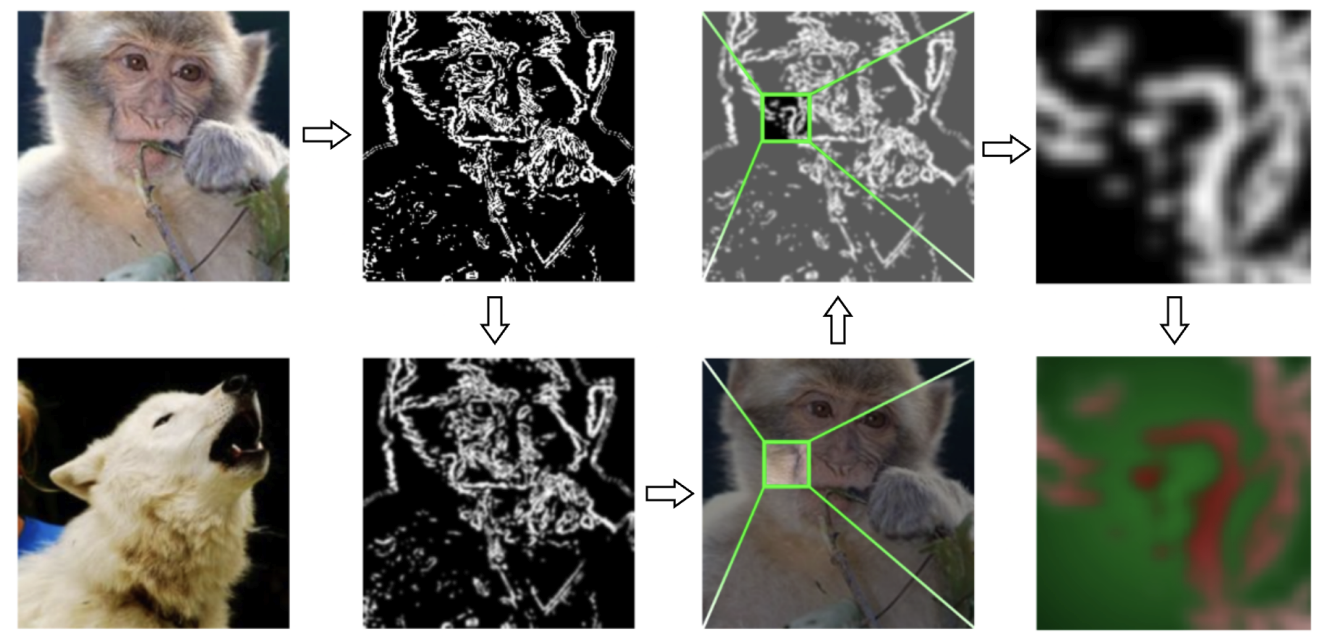
We propose Targeted Edge-informed Attack (TEA), a novel attack that utilizes edge information from the target image to carefully perturb it, thereby producing an adversarial image that is closer to the source image while still achieving the desired target classification. Our approach consistently outperforms current state-of-the-art methods across different models in low query settings (nearly 70% fewer queries are used), a scenario especially relevant in real-world applications with limited queries and black-box access. Furthermore, by efficiently generating a suitable adversarial example, TEA provides an improved target initialization for established geometry-based attacks.
A Swaminathan, M Akgün
Links:
SaTML 2026
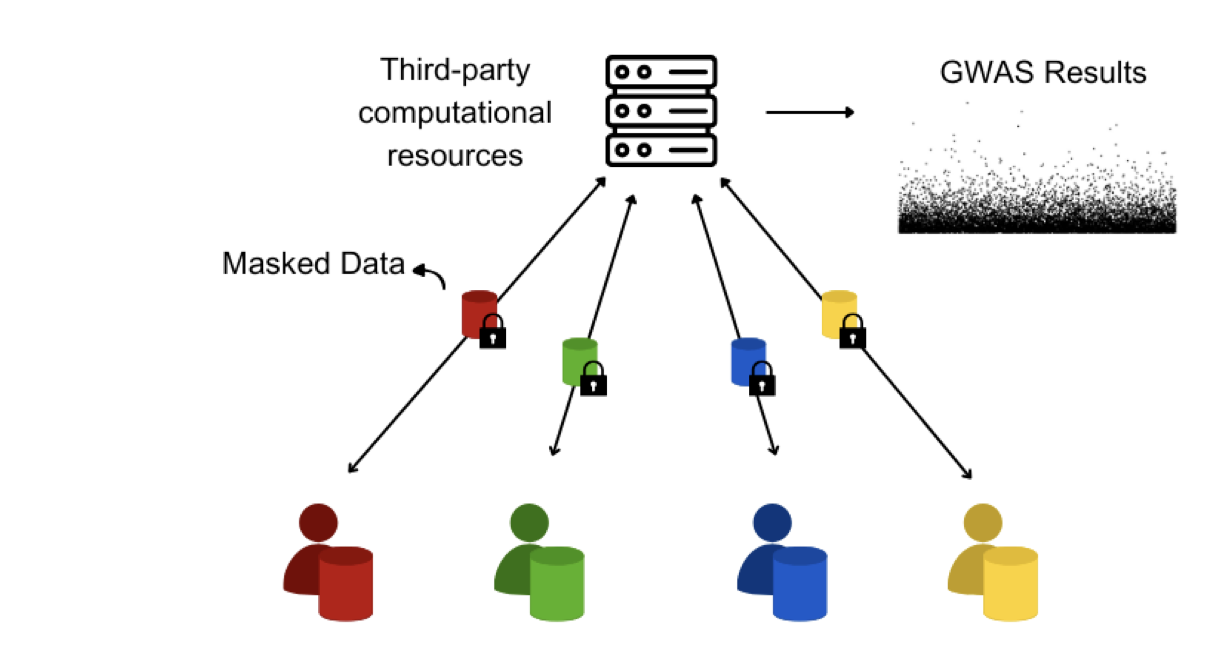
We present a novel algorithm PP-GWAS designed to improve upon existing standards in terms of computational efficiency and scalability without sacrificing data privacy. This algorithm employs randomized encoding within a distributed architecture to perform stacked ridge regression on a Linear Mixed Model to ensure rigorous analysis. Experimental evaluation with real world and synthetic data indicates that PP-GWAS can achieve computational speeds twice as fast as similar state-of-the-art algorithms while using lesser computational resources, all while adhering to a robust security model that caters to an all-but-one semi-honest adversary setting.
A Swaminathan, A Hannemann, AB Ünal, N Pfeifer, M Akgün
Links:
Nature Communications (2025)
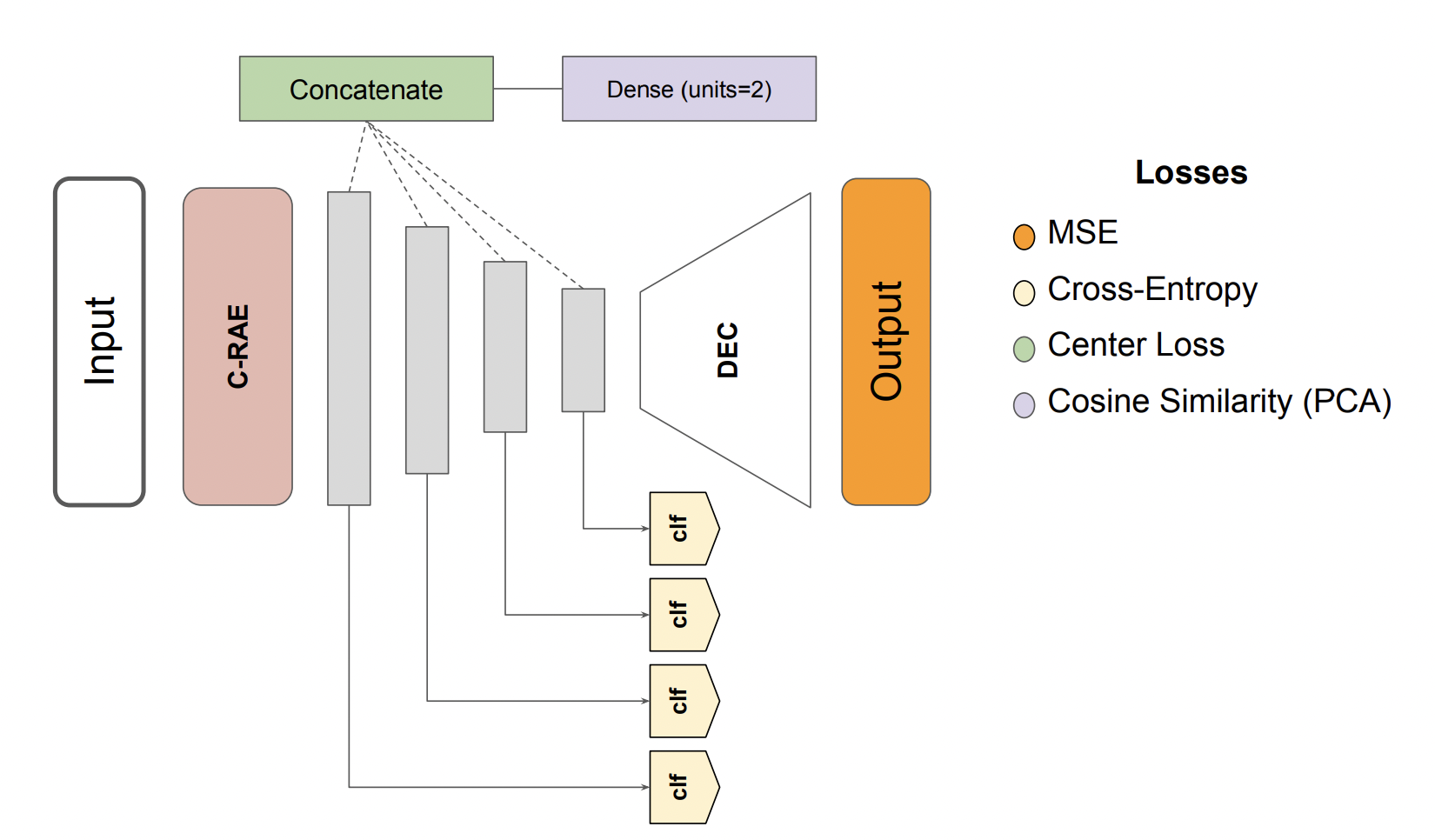
Several domains increasingly rely on machine learning in their applications. The resulting heavy dependence on data has led to the emergence of various laws and regulations around data ethics and privacy and growing awareness of the need for privacy-preserving machine learning (ppML). Current ppML techniques utilize methods that are either purely based on cryptography, such as homomorphic encryption, or that introduce noise into the input, such as differential privacy. The main criticism given to those techniques is the fact that they either are too slow or they trade off a model’s performance for improved confidentiality. To address this performance reduction, we aim to leverage robust representation learning as a way of encoding our data while optimising the privacy-utility trade-off.
S Ouaari, AB Ünal, M Akgün, N Pfeifer
Links:
IEEE Access (2025)
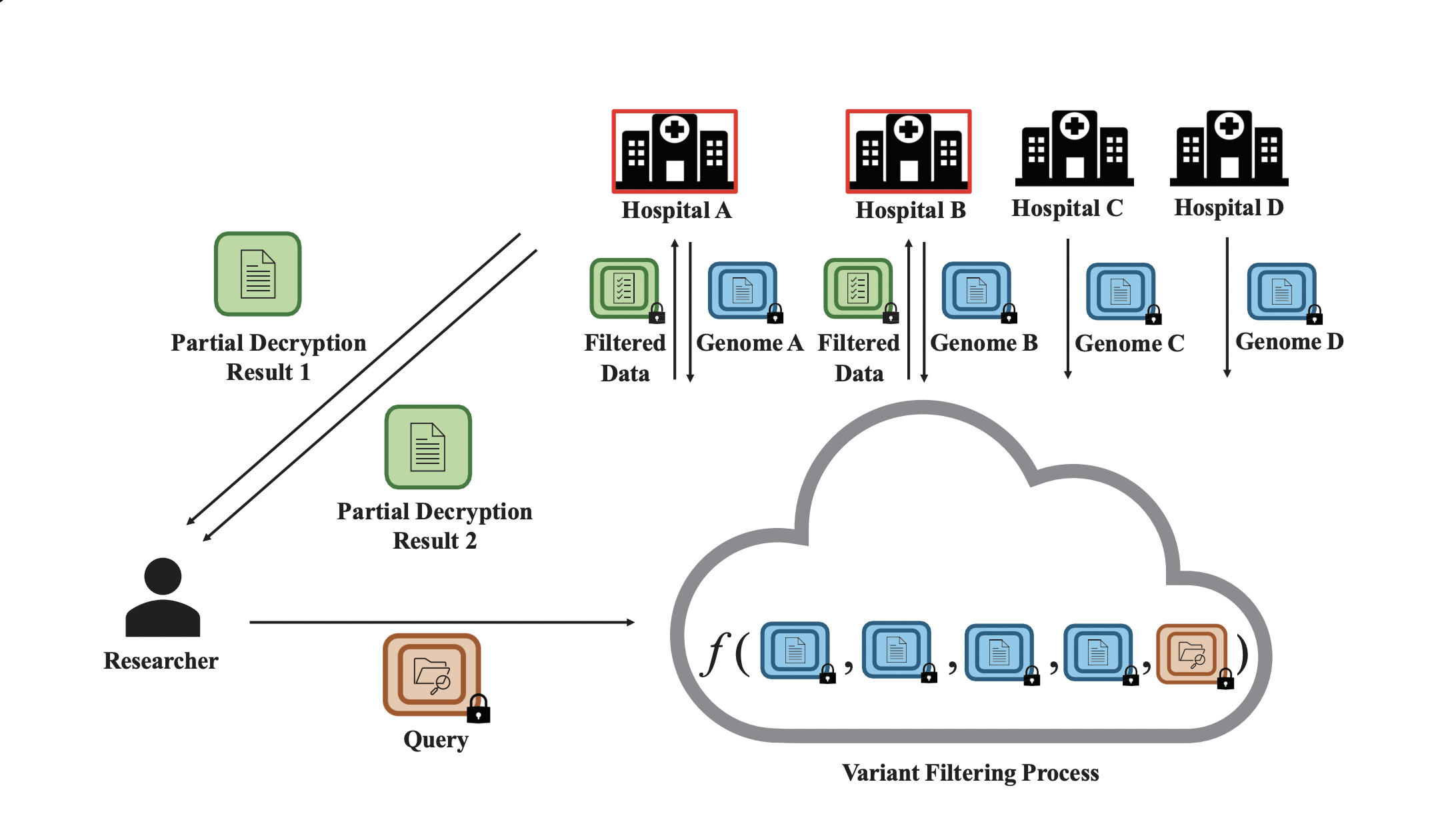
We present PRISM, a novel privacy-preserving framework based on fully homomorphic encryption (FHE) that facilitates rare disease variant analysis across multiple institutions without exposing sensitive genomic information. To address the challenges of centralized trust, PRISM is built upon a threshold FHE scheme. This approach decentralizes key management across participating institutions and ensures no single entity can unilaterally decrypt sensitive data. Our method filters disease-causing variants under recessive, dominant, and de novo inheritance models entirely on encrypted data.
G Akkaya, N Erdoğmuş, M Akgün
Links:
Bioinformatics (2025)
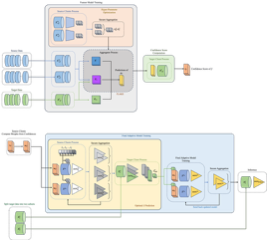
In this work, we propose freda, a privacy-preserving federated method for unsupervised domain adaptation in regression tasks. Unlike deep learning-based FDA approaches, freda is the first method to enable the federated training of Gaussian Processes to model complex feature relationships while ensuring complete data privacy through randomized encoding and secure aggregation. This allows for effective domain adaptation without direct access to raw data, making it wellsuited for applications involving high-dimensional, heterogeneous datasets.
CA Baykara, AB Ünal, N Pfeifer, M Akgün
Links:
Bioinformatics (2025)
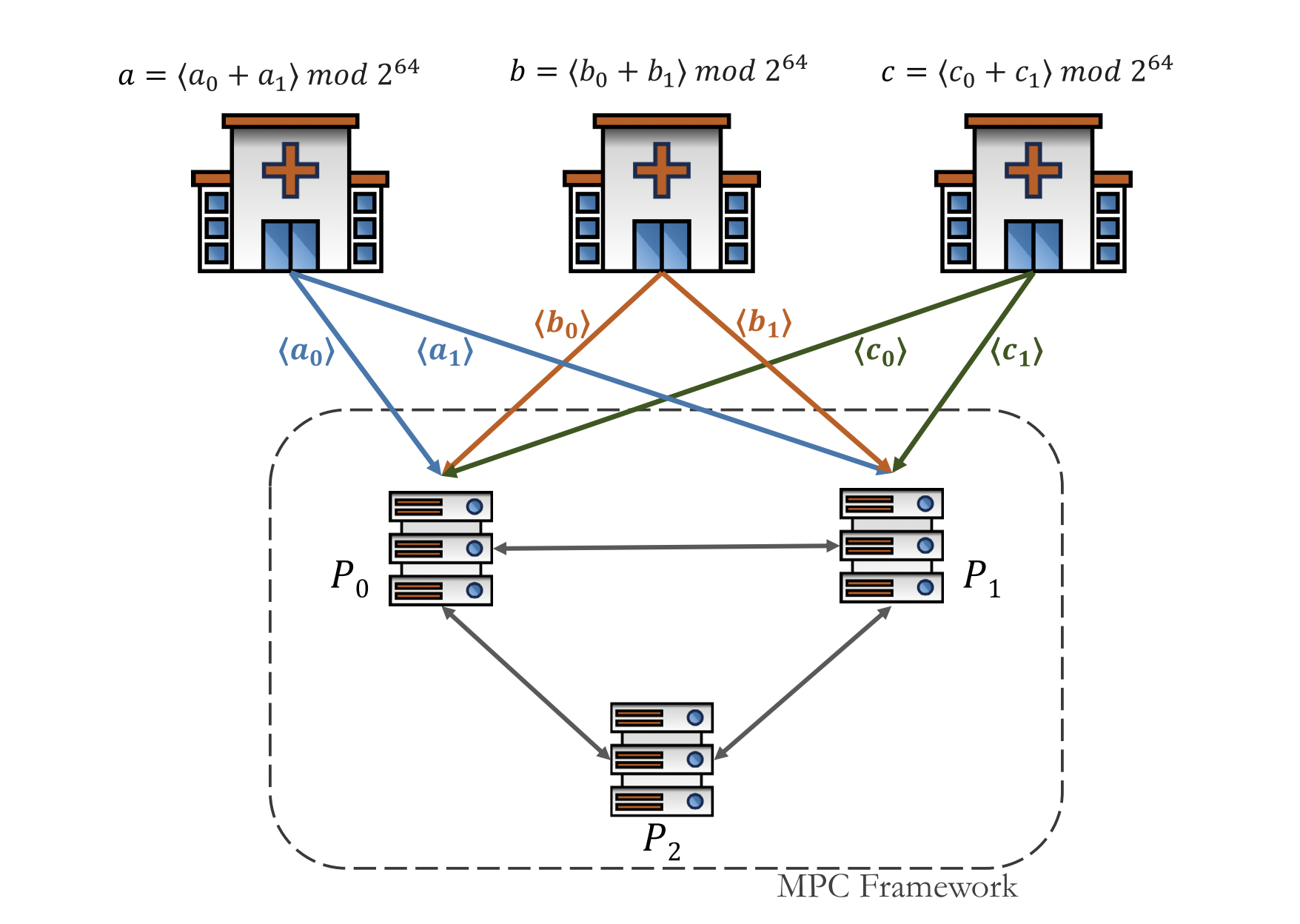
Machine learning models are being increasingly deployed in sensitive applications where data privacy and model security are of paramount importance. This paper introduces a novel privacy-preserving approach for logistic regression that integrates secret sharing-based multi-party computation (MPC) with differential privacy (DP). Our approach ensures input data confidentiality during training via a three-party MPC protocol that supports mini-batch gradient descent, reducing computational and communication overhead compared to prior MPC methods. To protect the model against membership inference and inversion attacks, we implement DP through direct gradient perturbation that scales efficiently to high-dimensional data while preserving strong (\epsilon)-differential privacy guarantees.
SS Magara, I Motorcu, ES Mektepli, CA Baykara, AB Ünal, M Akgün
Links:
IEEE Access (2025)
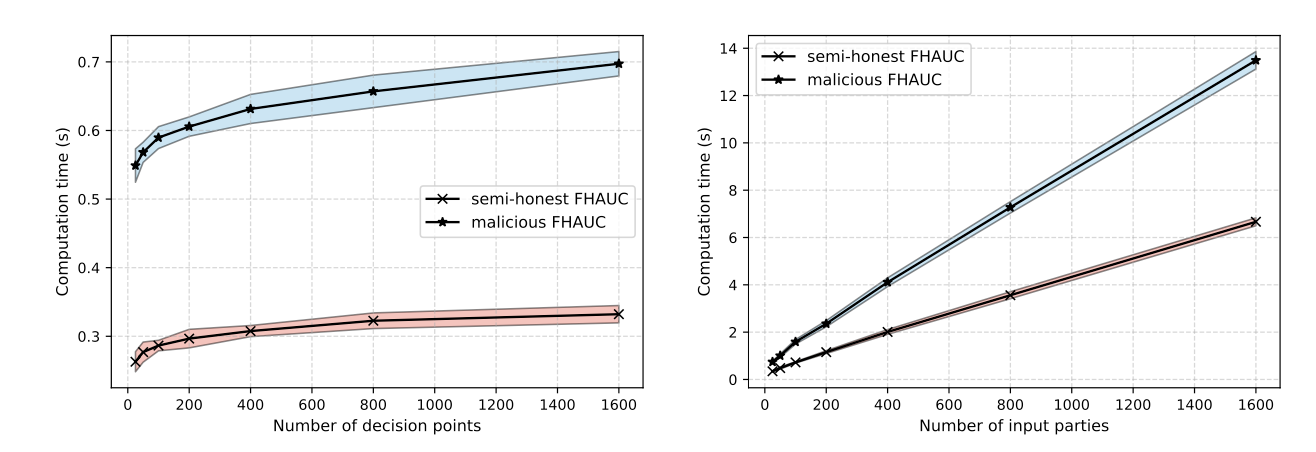
Current research on federated learning primarily focuses on preserving privacy during the training phase. However, model evaluation has not been adequately addressed, despite the potential for significant privacy leaks during this phase as well. In this paper, we demonstrate that the state-of-the-art AUC computation method for federated learning systems, which utilizes differential privacy, still leaks sensitive information about the test data while also requiring a trusted central entity to perform the computations.
CA Baykara, AB Ünal, M Akgün
Links:
IEEE TPS (2025)
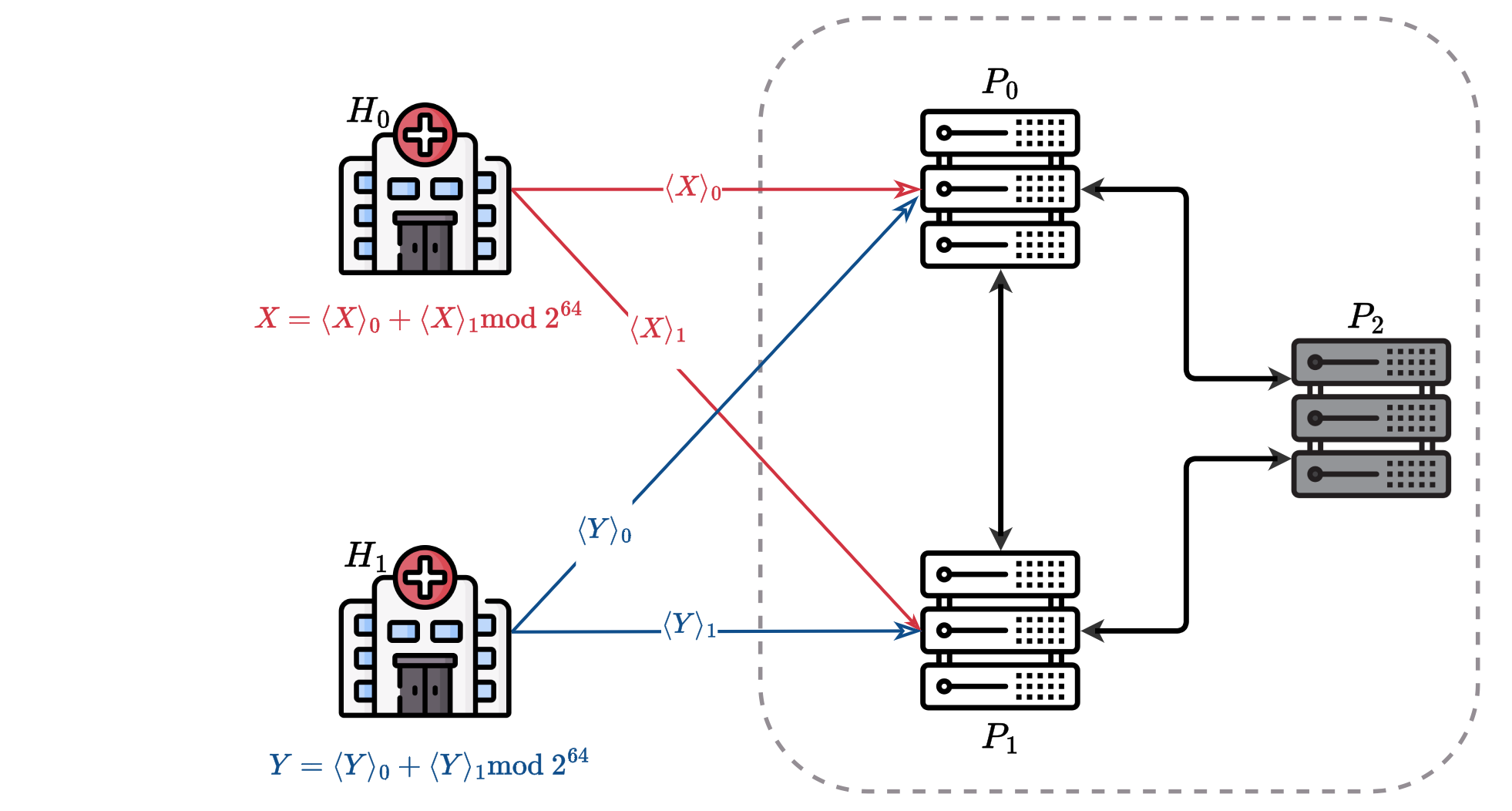
This work addresses the critical challenge of securely linking medical records across institutions while preserving patient privacy. Integrating data from various sources in healthcare can provide comprehensive insights into disease progression and treatment outcomes. However, strict privacy regulations often limit the ability to share this data. To tackle this, we propose a three-party Multi-Party Computation (MPC) Record Linkage method that ensures sensitive data remains confidential throughout the linkage process. By eliminating the need for Bloom filters and using bigram-based string similarity computations in a three-party framework, the method not only enhances privacy but also achieves an impressive up to 14x faster performance compared to state-of-the-art solutions. This scalable approach offers a practical solution for large-scale, privacy-preserving healthcare data integration.
ŞS Mağara, Noah Dietrich, AB Ünal, M Akgün
Links:
Journal of Biomedical Informatics (2025)
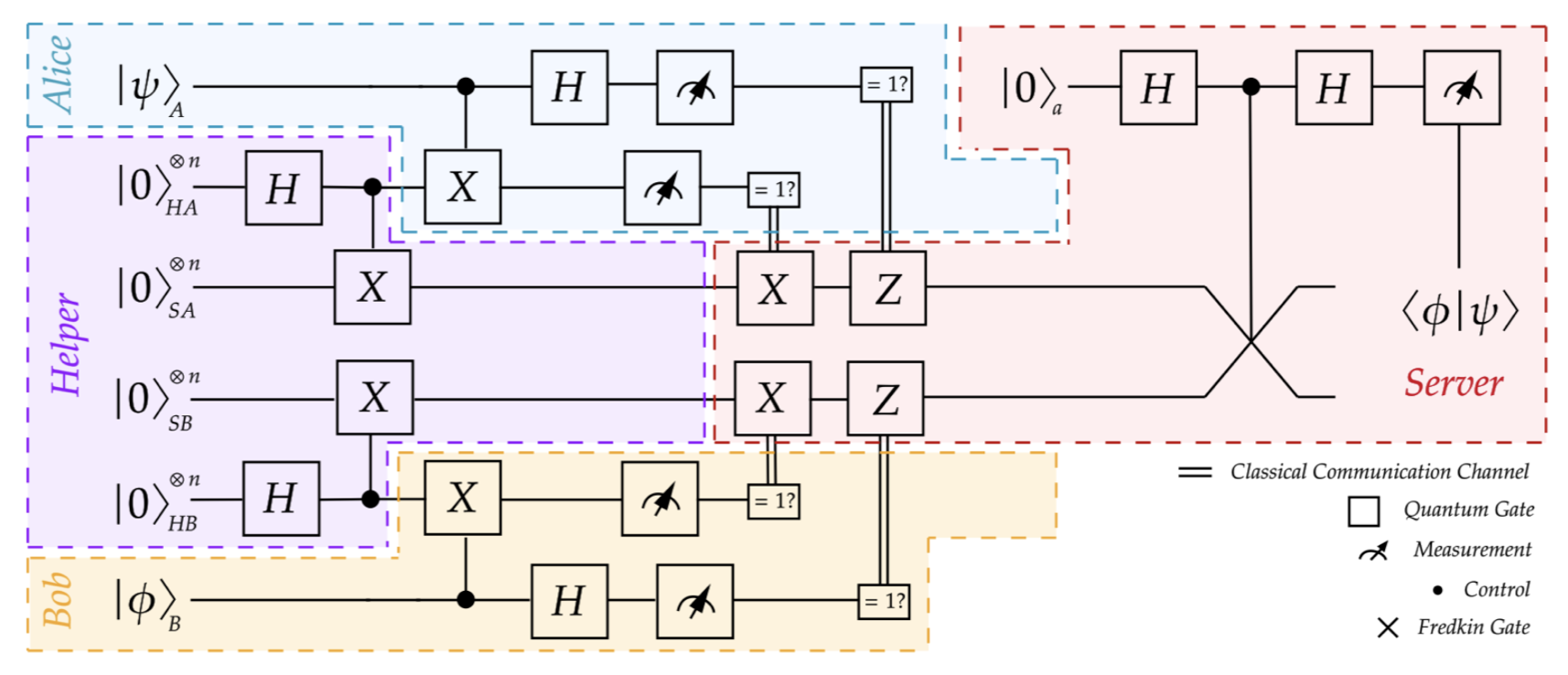
While advancements in secure quantum machine learning are notable, the development of secure and distributed quantum analogues of kernel-based machine learning techniques remains underexplored. In this work, we present a novel approach for securely computing common kernels, including polynomial, radial basis function (RBF), and Laplacian kernels, when data is distributed, using quantum feature maps.
A Swaminathan, M Akgün
Links:
TMLR
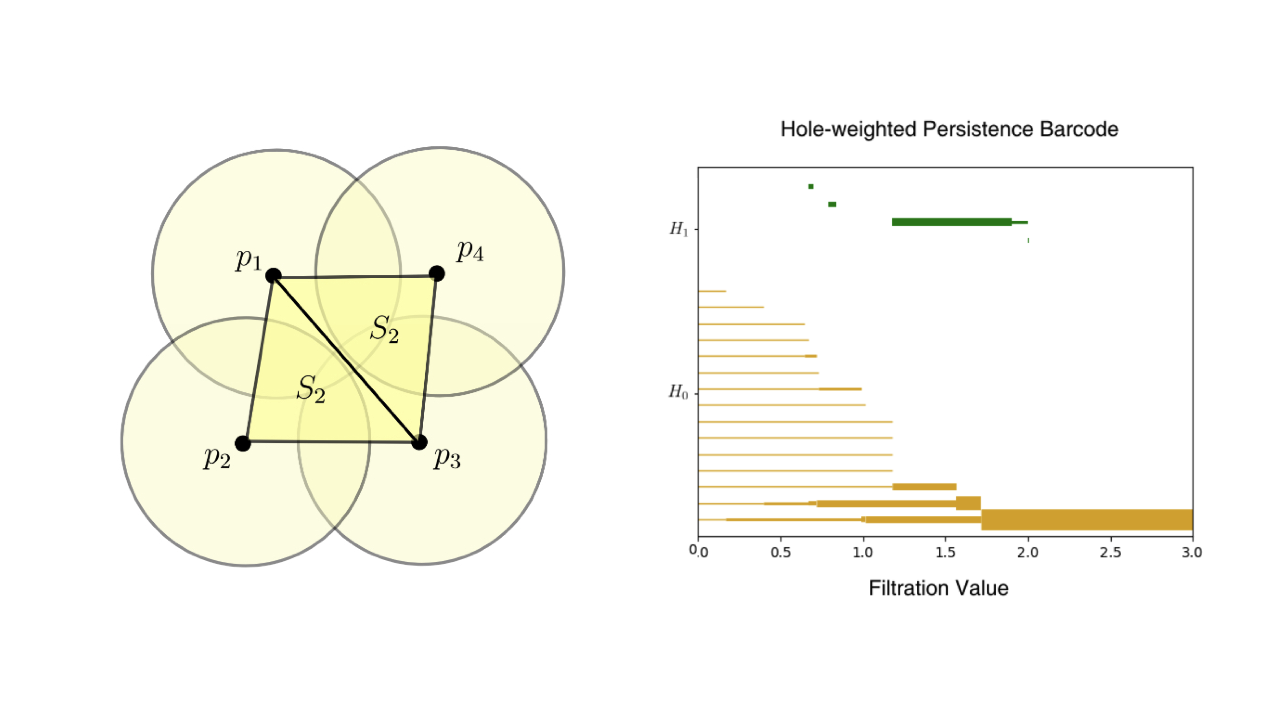
With the rapid digitization of Electronic Health Records (EHRs), fast and adaptive data anonymization methods have become crucial. While topological data analysis (TDA) tools have been proposed to anonymize static datasets—creating multiple generalizations for different anonymization needs from a single computation—their application to dynamic datasets remains unexplored. Our work adapts existing methodologies to dynamic settings by developing an improved version of weighted persistence barcodes that track higher-dimensional holes in data, allowing real-time editing of persistence information.
A Swaminathan, M Akgün
Links:
DPM 2024
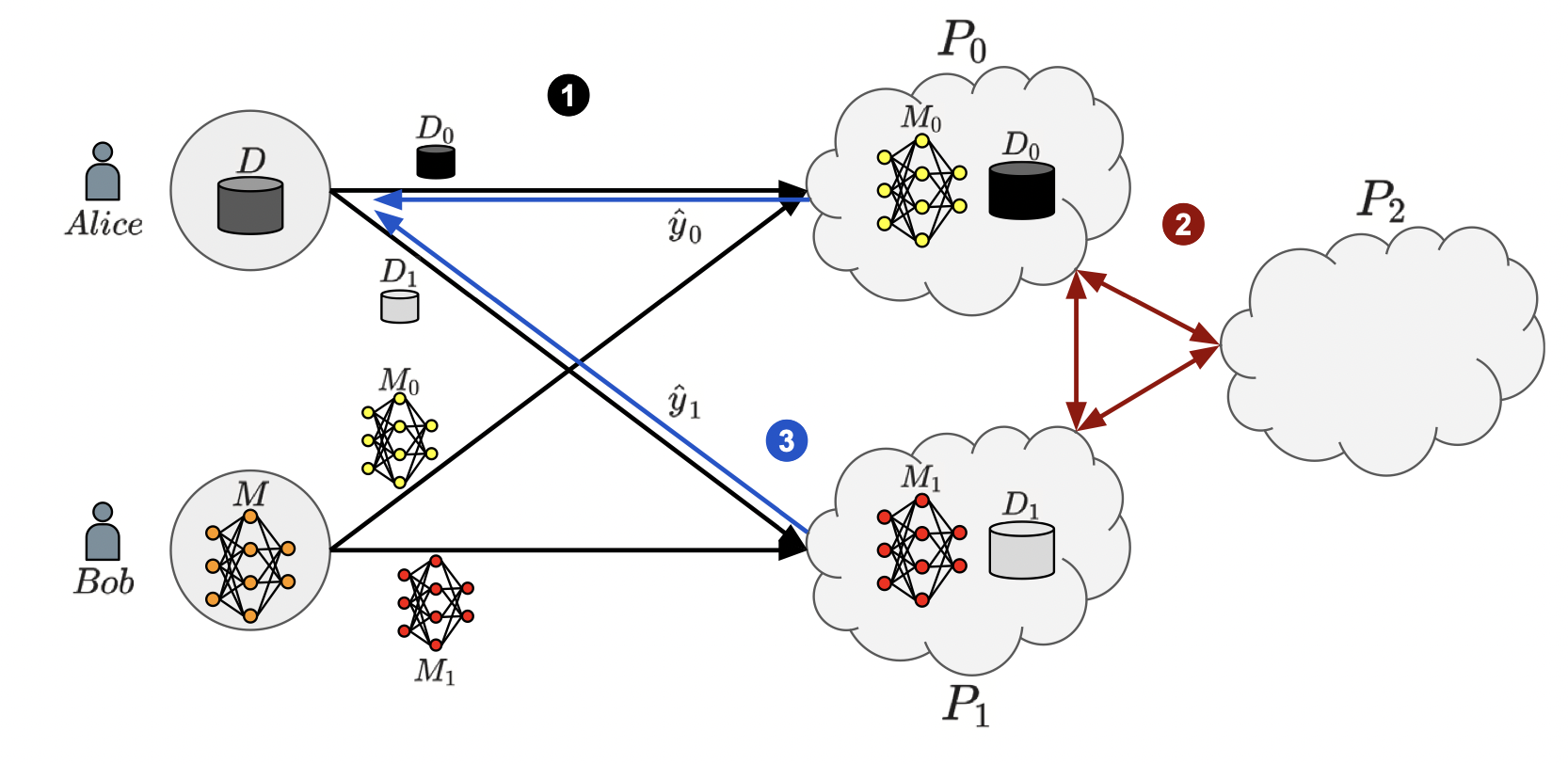
The complexity and cost of training machine learning models have made cloud-based machine learning as a service (MLaaS) attractive for businesses and researchers. MLaaS eliminates the need for in-house expertise by providing pre-built models and infrastructure. However, it raises data privacy and model security concerns, especially in medical fields like protein fold recognition. We propose a secure three-party computation-based MLaaS solution for privacy-preserving protein fold recognition, protecting both sequence and model privacy.
AB Ünal, N Pfeifer, M Akgün
Links:
Cell Patterns 101023 (2024)
GitHub Repository
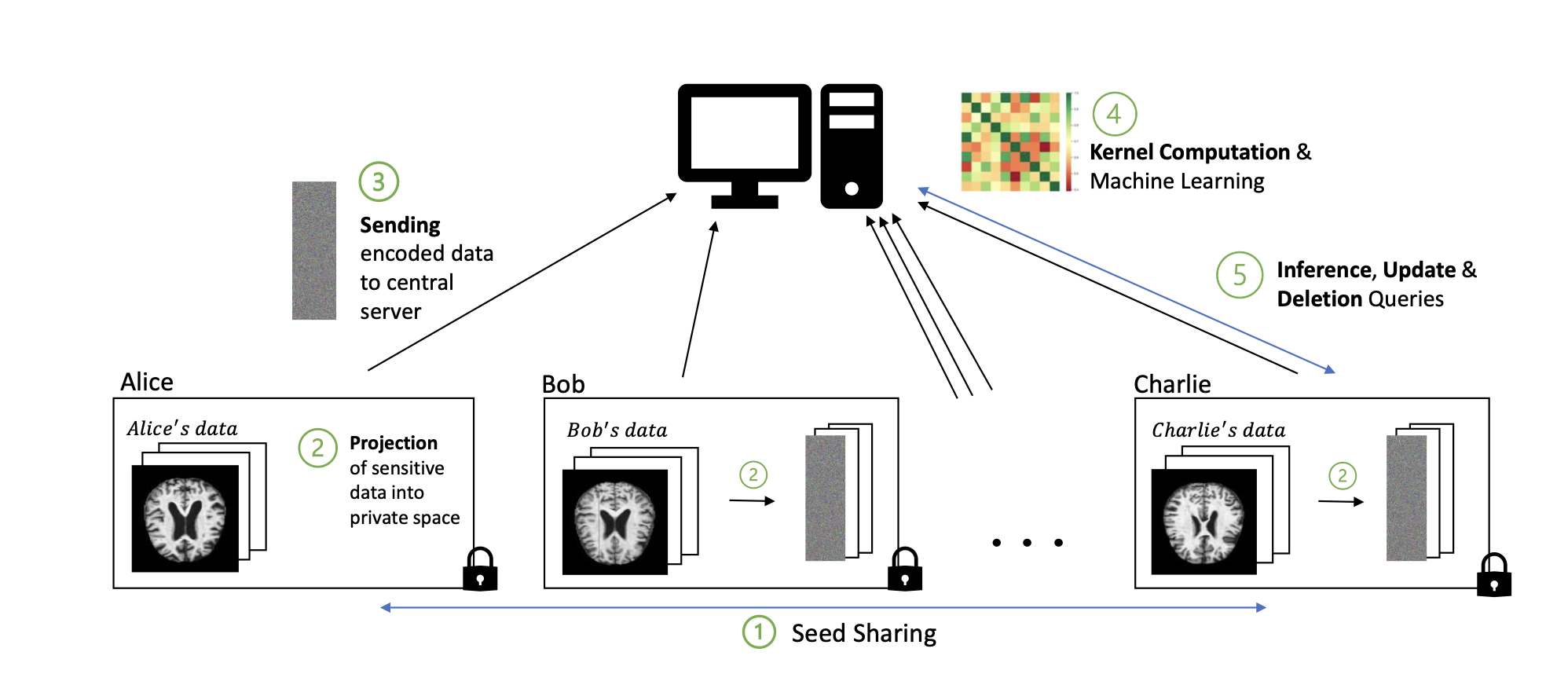
Addressing the need for efficient privacy- preserving methods on distributed image data, we introduce OKRA (Or- thonormal K-fRAmes), a novel randomized encoding-based approach for kernel-based machine learning. This technique, tailored for widely used kernel functions, significantly enhances scalability and speed compared to current state-of-the-art solutions. Through experiments conducted on various clinical image datasets, we evaluated model quality, computa- tional performance, and resource overhead. Additionally, our method outperforms comparable approaches.
A Hannemann, A Swaminathan, AB Ünal, M Akgün
Links:
CIBB (2024)
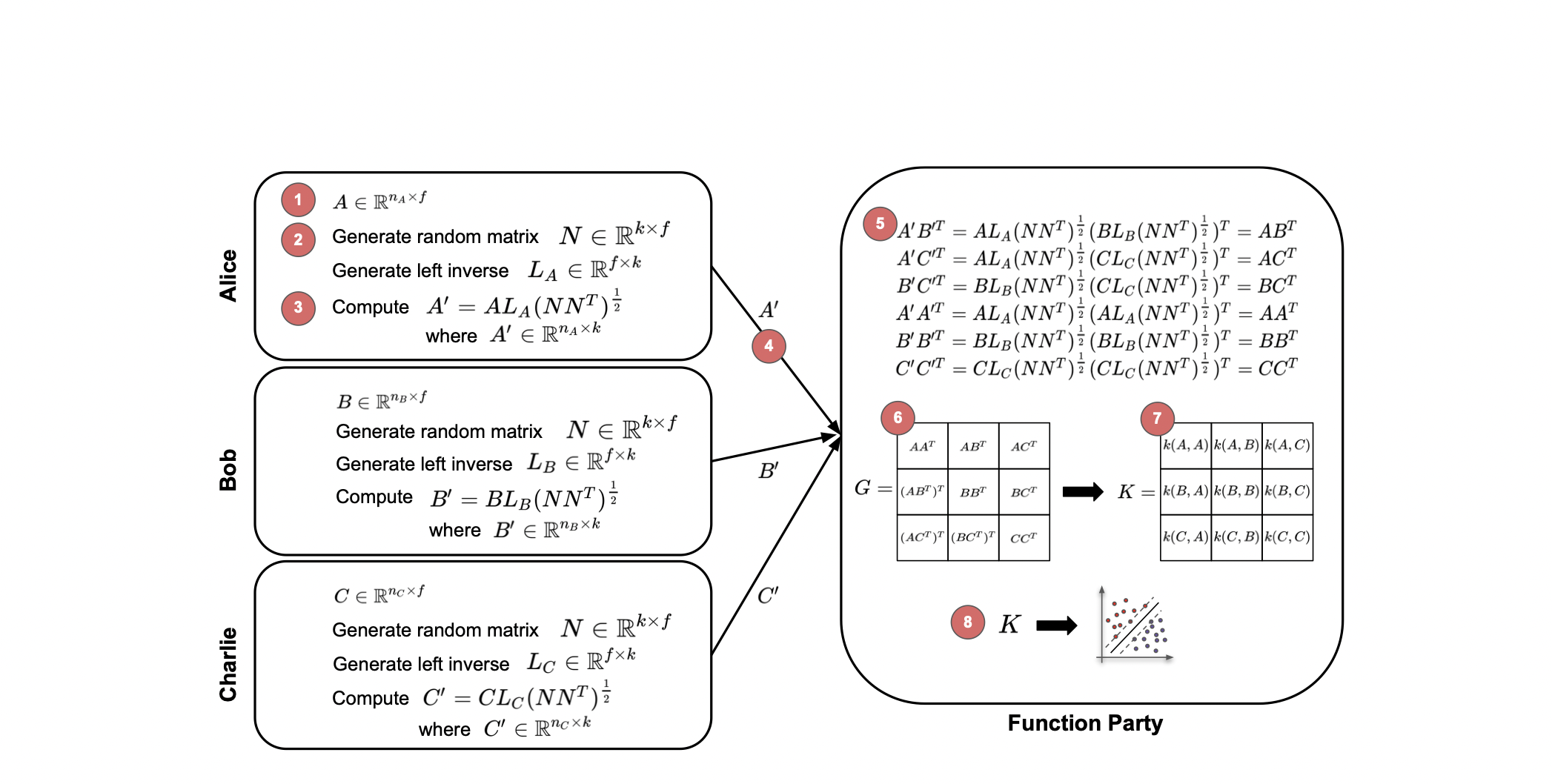
We’ve introduced FLAKE, a privacy-preserving Federated Learning Approach for Kernel methods on horizontally distributed data. By allowing data sources to mask their data, a centralized instance can generate a Gram matrix, preserving privacy while enabling the training of kernel-based algorithms like Support Vector Machines. We’ve established that FLAKE safeguards against semi-honest adversaries learning the input data or the number of features. Testing on clinical and synthetic data confirms FLAKE’s superior accuracy and efficiency over similar methods. Its data masking and Gram matrix computation times are significantly less than SVM training times, making it highly applicable across various use cases.
A Hannemann, AB Ünal, A Swaminathan, E Buchmann, M Akgün
Links:
IEEE TPS p82-90 (2023)
We propose a secure 3-party computation framework, CECILIA, offering PP building blocks to enable complex operations privately. In addition to the adapted and common operations like addition and multiplication, it offers multiplexer, most significant bit and modulus conversion. The first two are novel in terms of methodology and the last one is novel in terms of both functionality and methodology. CECILIA also has two complex novel methods, which are the exact exponential of a public base raised to the power of a secret value and the inverse square root of a secret Gram matrix.
AB Ünal, N Pfeifer, M Akgün
Links:
arXiv:2202.03023 (2022)
Disease–gene association studies are of great importance. However, genomic data are very sensitive when compared to other data types and contains information about individuals and their relatives. We propose a method that uses secure multi-party computation to query genomic databases in a privacy-protected manner.
M Akgün, N Pfeifer, O Kohlbacher
Links:
Bioinformatics 38,8 (2022)
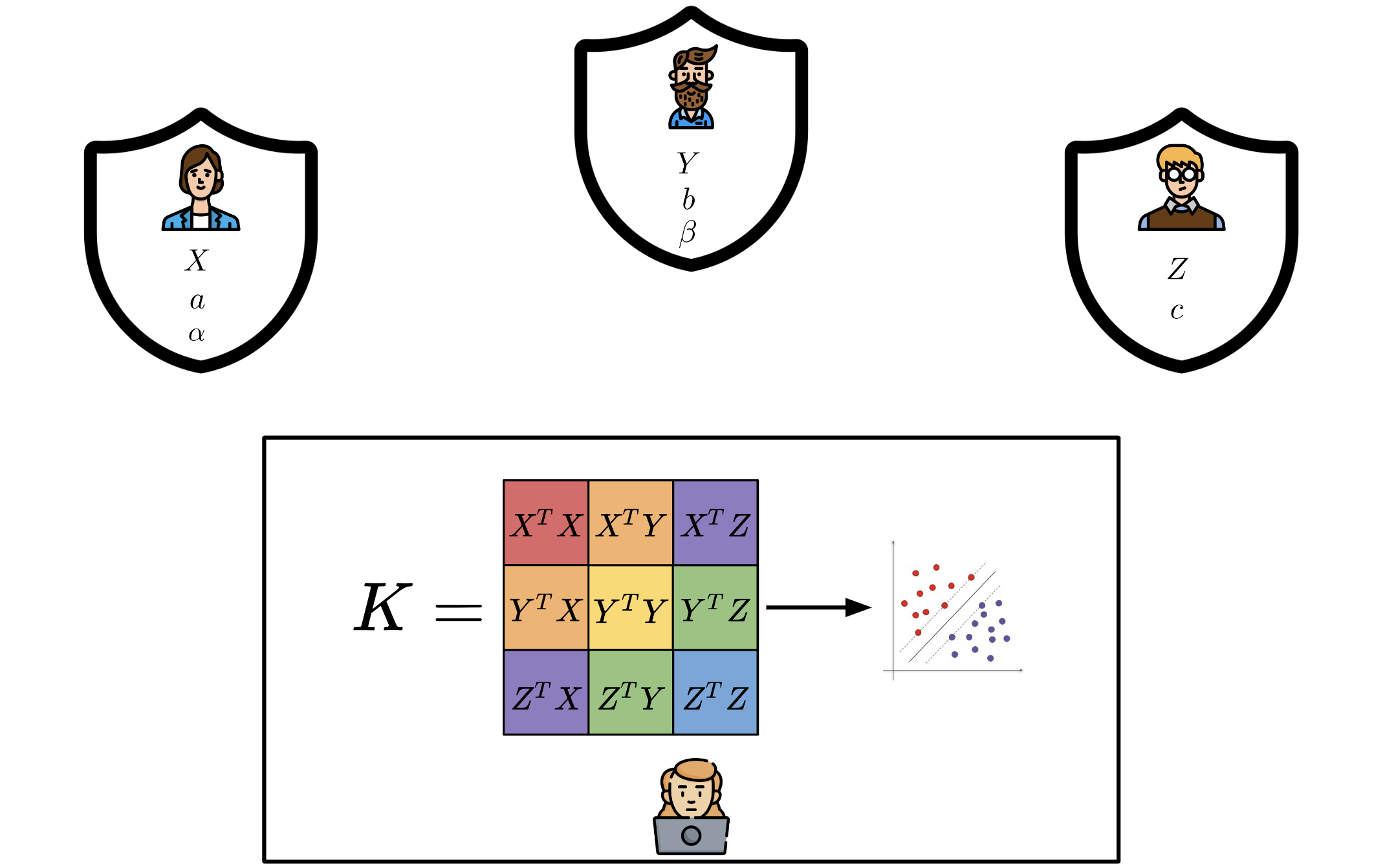
We introduce ESCAPED, which stands for Efficient SeCure And PrivatE Dot product framework. ESCAPED enables the computation of the dot product of vectors from multiple sources on a third-party, which later trains kernel-based machine learning algorithms, while neither sacrificing privacy nor adding noise.
AB Ünal, M Akgün, N Pfeifer
Links:
AAAI 35, 11 (2021)
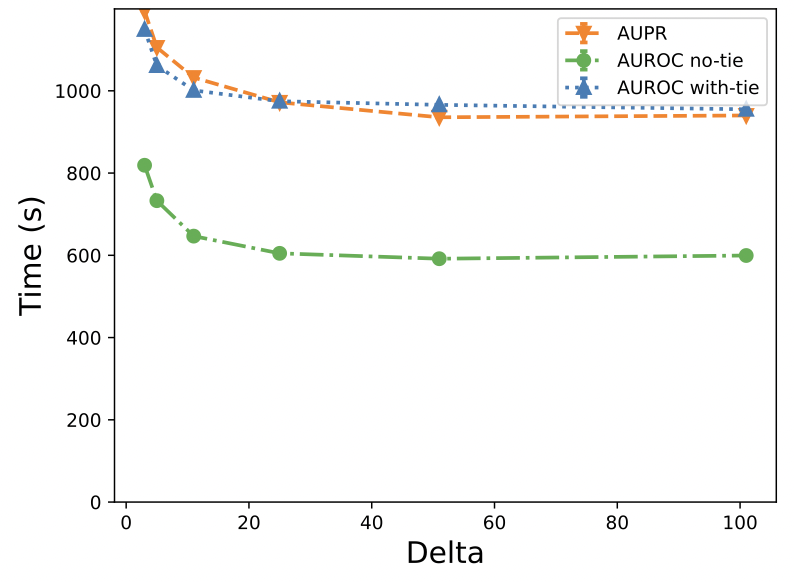
In this paper, we propose an MPC-based framework, called PPAURORA with private merging of sorted lists and novel methods for comparing two secret-shared values, selecting between two secret-shared values, converting the modulus, and performing division to compute the exact AUC as one could obtain on the pooled original test samples.
AB Ünal, N Pfeifer, M Akgün
Links:
arXiv:2102.08788 (2021)
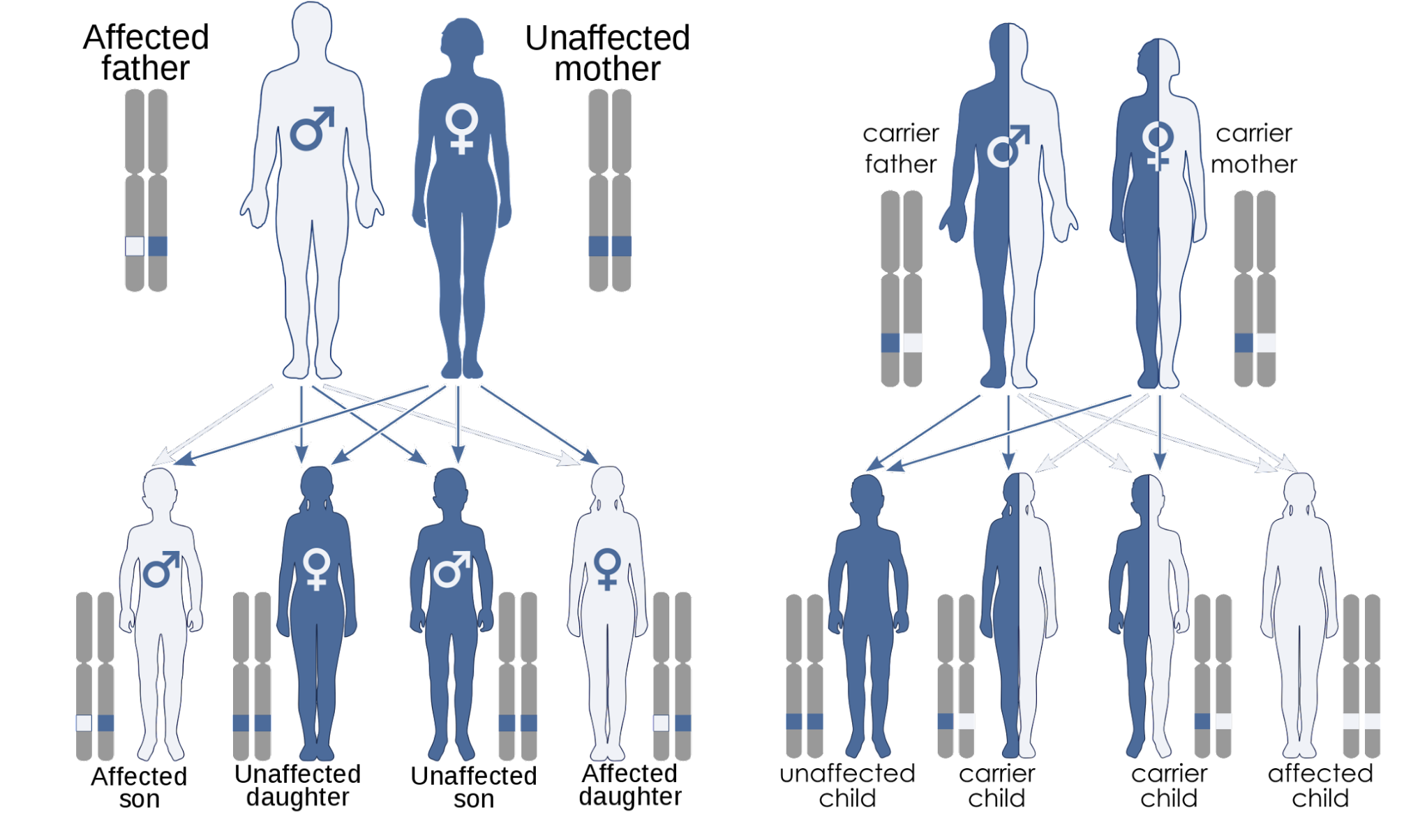
We present an approach to identify disease-associated variants and genes while ensuring patient privacy. The proposed method uses secure multi-party computation to find disease-causing mutations under specific inheritance models without sacrificing the privacy of individuals. It discloses only variants or genes obtained as a result of the analysis. Thus, the vast majority of patient data can be kept private.
M Akgün, AB Ünal, B Ergüner, N Pfeifer, O Kohlbacher
Links:
Bioinformatics 36,21 (2021)